11.1.2. Macaque template: NMT v2 (current)¶
Overview¶
Here we present the NIMH Macaque Template (NMT), version 2.0. This is a group template from 31 macaques with surfaces and a 5-class tissue segmentation.
The NMT v2 is the most current version of this dataset.
Be sure to also check out the accompanying CHARM (Cortical Hierarchy Atlas of the Rhesus Macaque) dataset, described here.
Contents¶
There are both symmetric and asymmetric “variants” for the NMT v2.
Additionally, for each variant there are sets of data with different spatial resolution and FOV:
the “standard” NMT has 0.25 mm isotropic voxels and a “brain-focused” FOV,
the “low-res” NMT has 0.5 mm isotropic voxels and a “brain-focused” FOV,
the “full-head” NMT has 0.25 mm isotropic voxels and a larger FOV, encompassing more non-brain material.
An example of the contents in the download of the symmetric NMT v2 (which is essentially mirrored in the asymmetric version) is provided here:
NMT v2 contents |
Description |
|---|---|
NMT_v2.0_sym/ |
directory of the “standard” NMT files (i.e., standard spatial resolution of 0.25 mm iso voxels and brain-focused FOV) |
NMT_v2.0_sym_05mm/ |
directory of the “low-res” NMT files (0.5 mm iso voxels; lower spatial resolution but still useful, if not preferred, for some analysis cases like in standard FMRI processing) |
NMT_v2.0_sym_fh/ |
directory of the “full head” NMT files (i.e., 0.25 mm iso voxels but a larger FOV, more non-brain material), which may be useful for some alignment cases |
NMT_v2.0_sym_surfaces/ |
directory of GIFTI surfaces of many of the tissue/segmentation regions |
CustomAtlases.niml |
atlas description text file for AFNI |
NMT_changelog.txt |
text file of changes and updates to the distributed datasets |
NMT_v2.0_sym_env.csh |
script to set AFNI environment variables for AFNI to use
atlases and make AFNI more monkey friendly. Note: running
this script will change your default atlases for |
supplemental_CHARM/ |
directory of tables of CHARM ROI index/label definitions and more |
supplemental_D99/ |
directory of a table of D99 ROI index/label definitions |
An example of the contents of the “standard” NMT directory NMT_v2.0_sym/, above are as follows (within similar sets of data in the “low-res” and “full-head” directories):
NMT_v2.0_sym/ contents |
Description |
|---|---|
CHARM_in_NMT_v2.0_sym.nii.gz |
hierarchical atlas (6 levels) in NMT v2 space |
D99_atlas_in_NMT_v2.0_sym.nii.gz |
D99 atlas in NMT v2 space |
NMT_v2.0_sym_brainmask.nii.gz |
binary brain mask (including cerebellum) |
NMT_v2.0_sym_GM_cortical_mask.nii.gz |
binary mask of cortical GM |
NMT_v2.0_sym.nii.gz |
the NMT v2 template, with non-brain material surrounding |
NMT_v2.0_sym_segmentation.nii.gz |
tissue segmentation of the whole brain |
NMT_v2.0_sym_SS.nii.gz |
the NMT itself, skullstripped, not stirred |
supplemental_CHARM/ |
the CHARM atlas levels as individual volumetric files |
supplemental_masks/ |
masks of “other” things (cerebellum, ventricles, L-R hemispheres) |
Example images¶
Full NMT with brainmask |
|---|
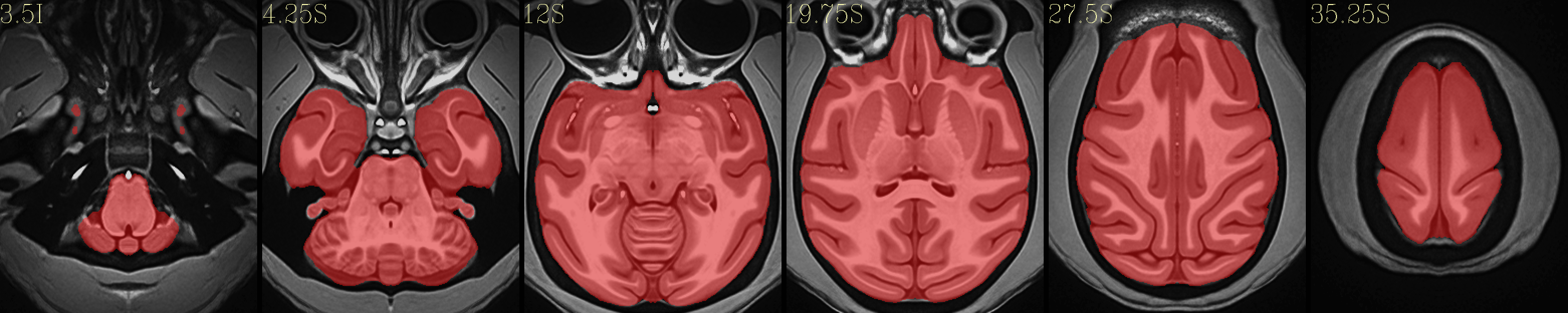
|
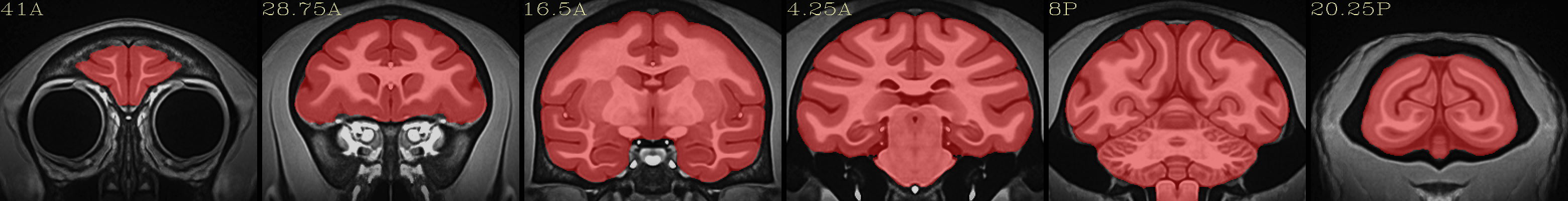
|
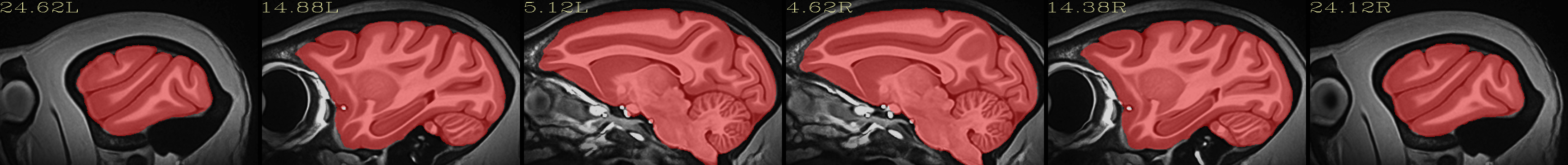
|
Skull-stripped NMT with 5-tissue segmentation |
|---|
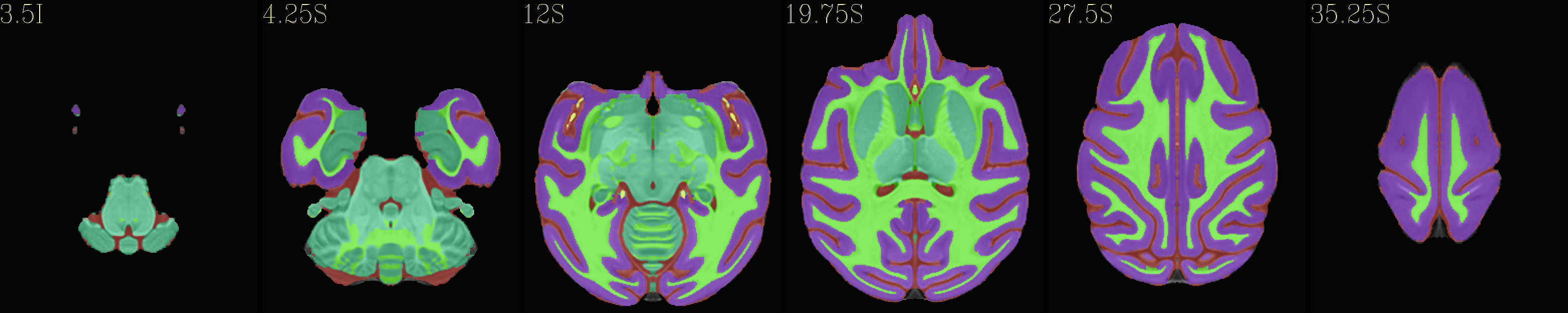
|
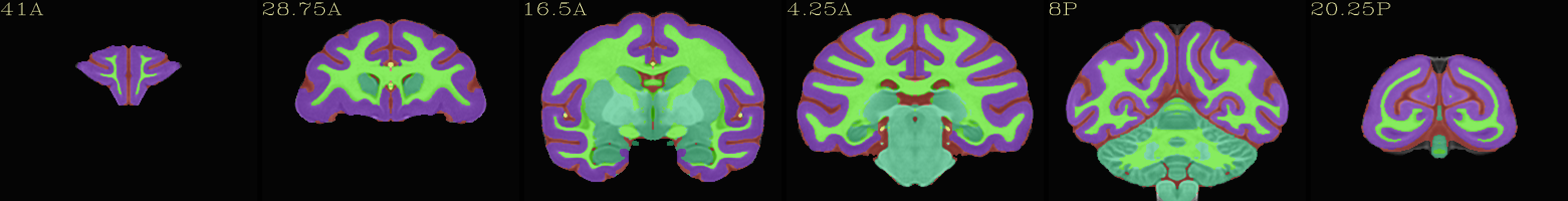
|
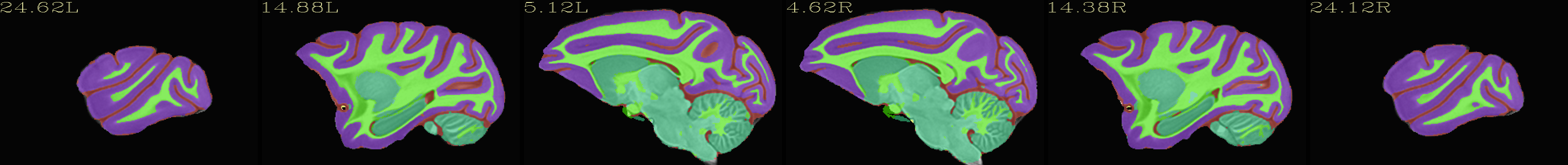
|
Skull-stripped NMT with the CHARM (level 2) |
|---|
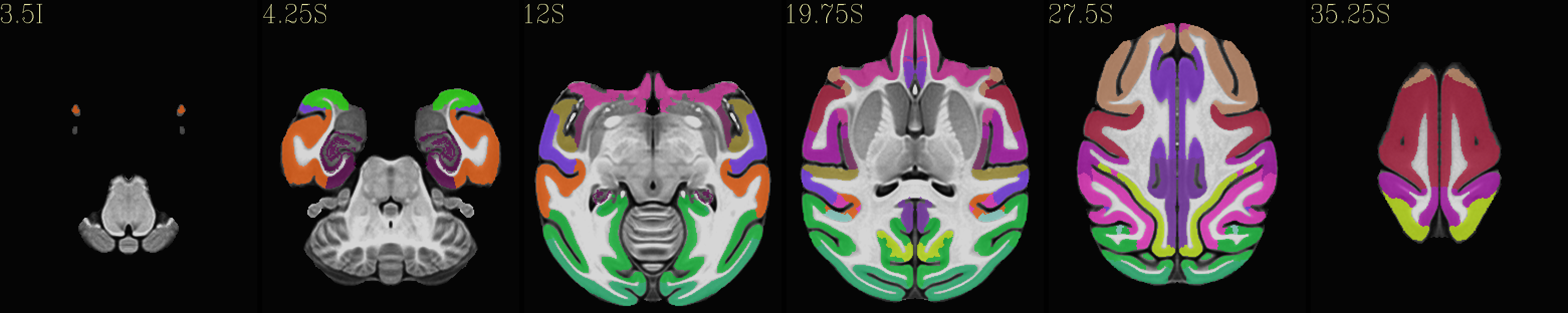
|

|
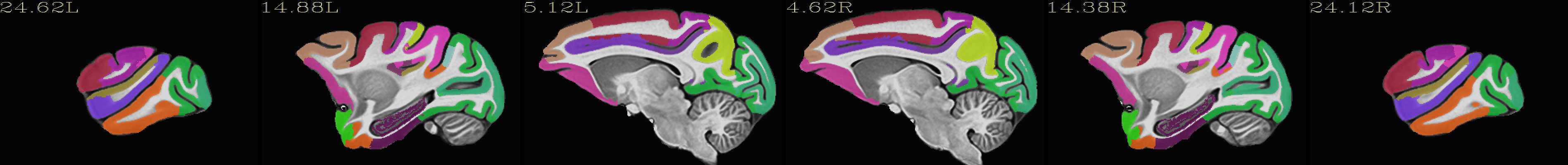
|
Skull-stripped NMT with the CHARM (level 5) |
|---|
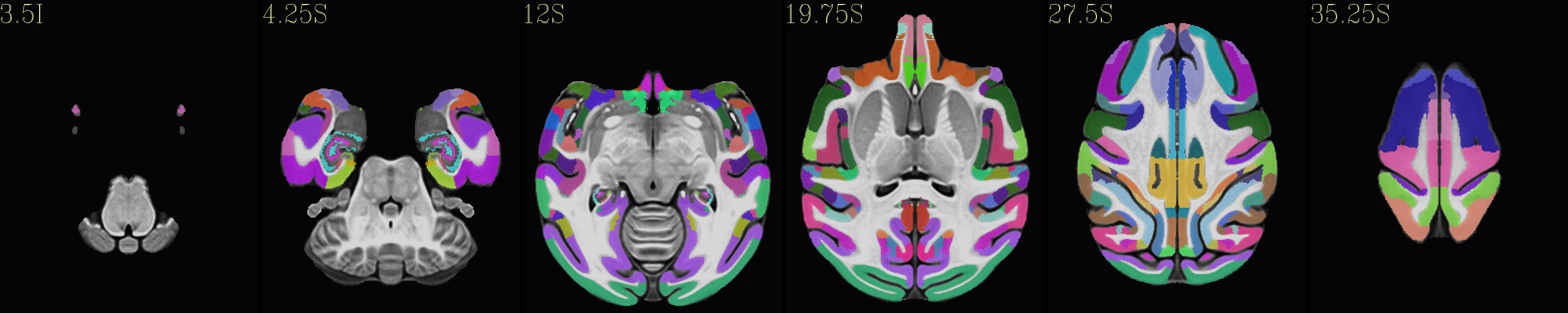
|
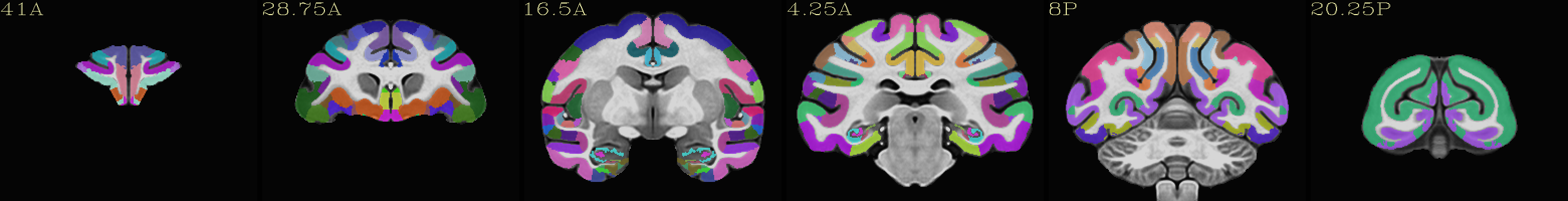
|
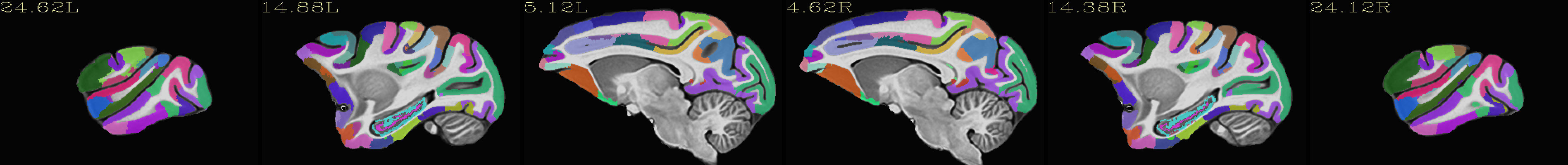
|
The script used to make these images with ``@chauffeur_afni`` is
available here: do_view_nmt_v2.0_sym.tcsh.
Download symmetric NMT v2 datasets¶
You can download and unpack the symmetric variant/form of the NMT v2 in any of the following ways:
(the AFNI way) copy+paste:
@Install_NMT -nmt_ver 2.0 -sym sym
(the plain Linux-y terminal way) copy+paste:
curl -O https://afni.nimh.nih.gov/pub/dist/atlases/macaque/nmt/NMT_v2.0_sym.tgz tar -xvf NMT_v2.0_sym.tgz
- (the mouseclick+ way) click on this link,... and then unpack the zipped directory by either clicking on it or using the above
tarcommand.
Download asymmetric NMT v2 datasets¶
You can download and unpack the asymmetric variant/form of the NMT v2 in any of the following ways:
(the AFNI way) copy+paste:
@Install_NMT -nmt_ver 2.0 -sym asym
(the Linux-y terminal way) copy+paste:
curl -O https://afni.nimh.nih.gov/pub/dist/atlases/macaque/nmt/NMT_v2.0_asym.tgz tar -xvf NMT_v2.0_asym.tgz
- (the mouseclick+ way) click on this link,... and then unpack the zipped directory by either clicking on it or using the above
tarcommand.
Citation/questions¶
If you make use of the NMT v2 template or accompanying data in your research, please cite:
Jung B, Taylor PA, Seidlitz PA, Sponheim C, Glen DR, Messinger A (2020). “A Comprehensive Macaque FMRI Pipeline and Hierarchical Atlas.” NeuroImage, submitted.Seidlitz J, Sponheim C, Glen DR, Ye FQ, Saleem KS, Leopold DA, Ungerleider L, Messinger A (2018). “A Population MRI Brain Template and Analysis Tools for the Macaque.” NeuroImage 170: 121–31.
