12.17.1. SurfLayers Demo 1¶
Introduction¶
We briefly describe the SurfLayers Demo, which combines AFNI+SUMA functionality to investigate surface layers in MRI data. This Demo was presented at OHBM 2021:
Creating Layered Surfaces to Visualize with AFNI + SUMA, with applications to laminar fMRIby Torrisi S, Lauren P, Taylor PA, Park S, Feinberg D, Glen DR.
To run this demo, your system’s AFNI version should be at least 21.1.14.
The Demo of scripts+data can be downloaded by running:
@Install_SURFLAYERS_DEMO1
The above command will both download and unpack the SurfLayers Demo
within the directory from which it is run. There is a descriptive
README.txt for users to ignore read.
This demo makes use of and presents several new (at the time) features
within AFNI and SUMA. Firstly, these scripts utilize the
SurfLayers and quickspecSL programs, to estimate intermediate
surface layers. Secondly, they utilize the clipping plane mode
within SUMA, whereby one can focus on subregions within the viewer and
investigate nested surface layers.
Each script uses terminal commands to “drive” both the afni and
suma GUIs, so you can visualize your data. That is in fact one of
the purposes of these new tools (and of AFNI in general): to keep
users close to their data, to understand it better. The GUIs also
“talk” to one another, sending information back and forth in realtime,
so you can really explore your data in fun and informative ways.
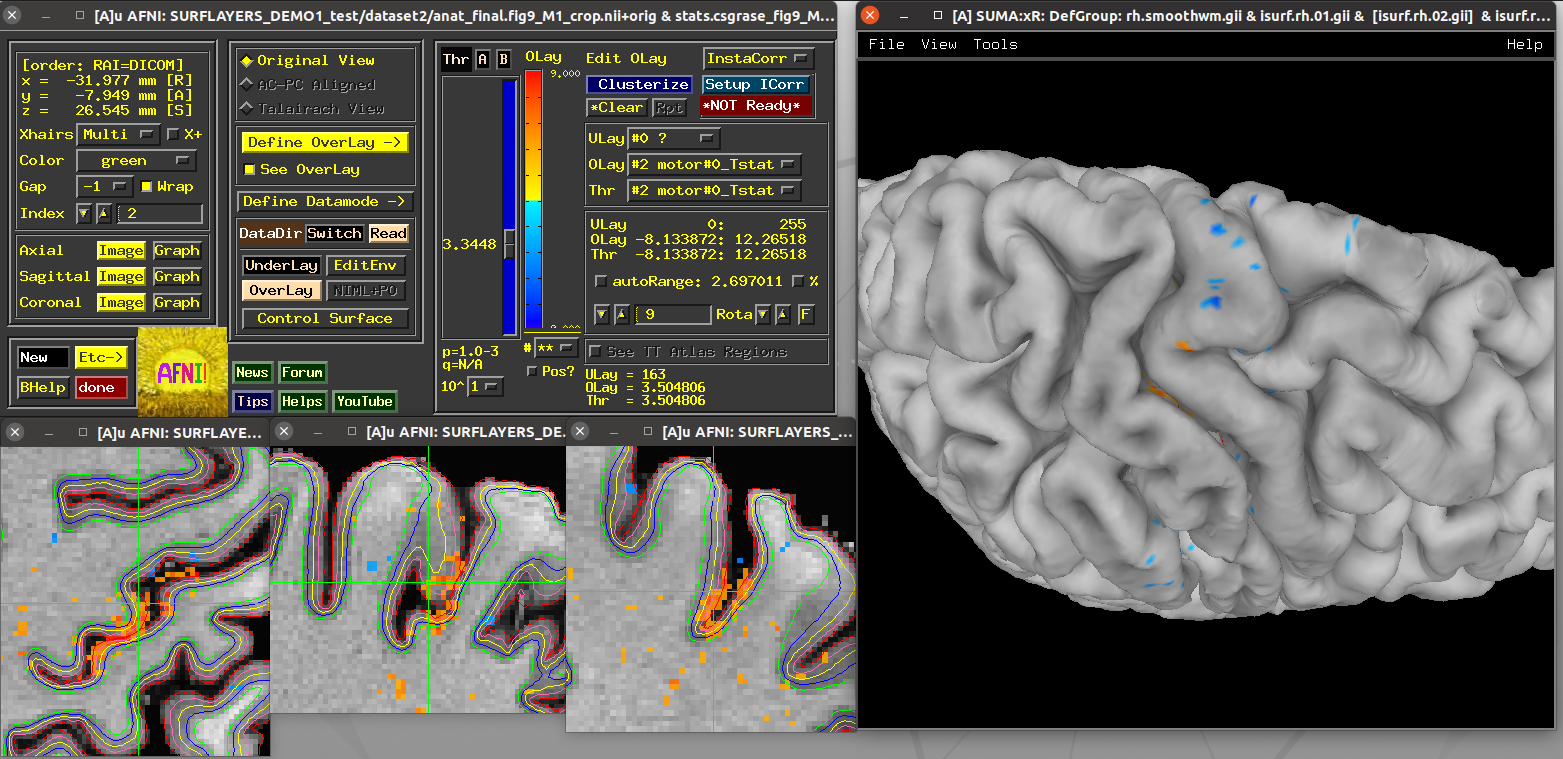
Scripts 1 and 2: basic MRI surface¶
These scripts use data from the AFNI Bootcamp, namely within the
AFNI_data6/FT_analyis/ directory (which is part of the standard
CD.tgz Bootcamp data package):
the
FT/SUMA/*data, which were created by running FreeSurfer’srecon-allon the subject’s anatomical, followed by AFNI’s@SUMA_Make_Spec_FSto create NIFTI volumes and standardized surface meshes.the results directory made by running the
s05.ap.uberscript, which contains anafni_proc.pycommand. (One could also runs05.ap.uber.NLthere, which runs anafni_proc.pycommand that makes using on nonlinear alignment between the subject and template space, for more detailed matching).
These scripts are not really doing laminar work, per se, because this data isn’t high enough resolution. However, these scripts do give a good intro to the functionality and viewing, at a resolution many people are used to working.
Running script |
|---|
|
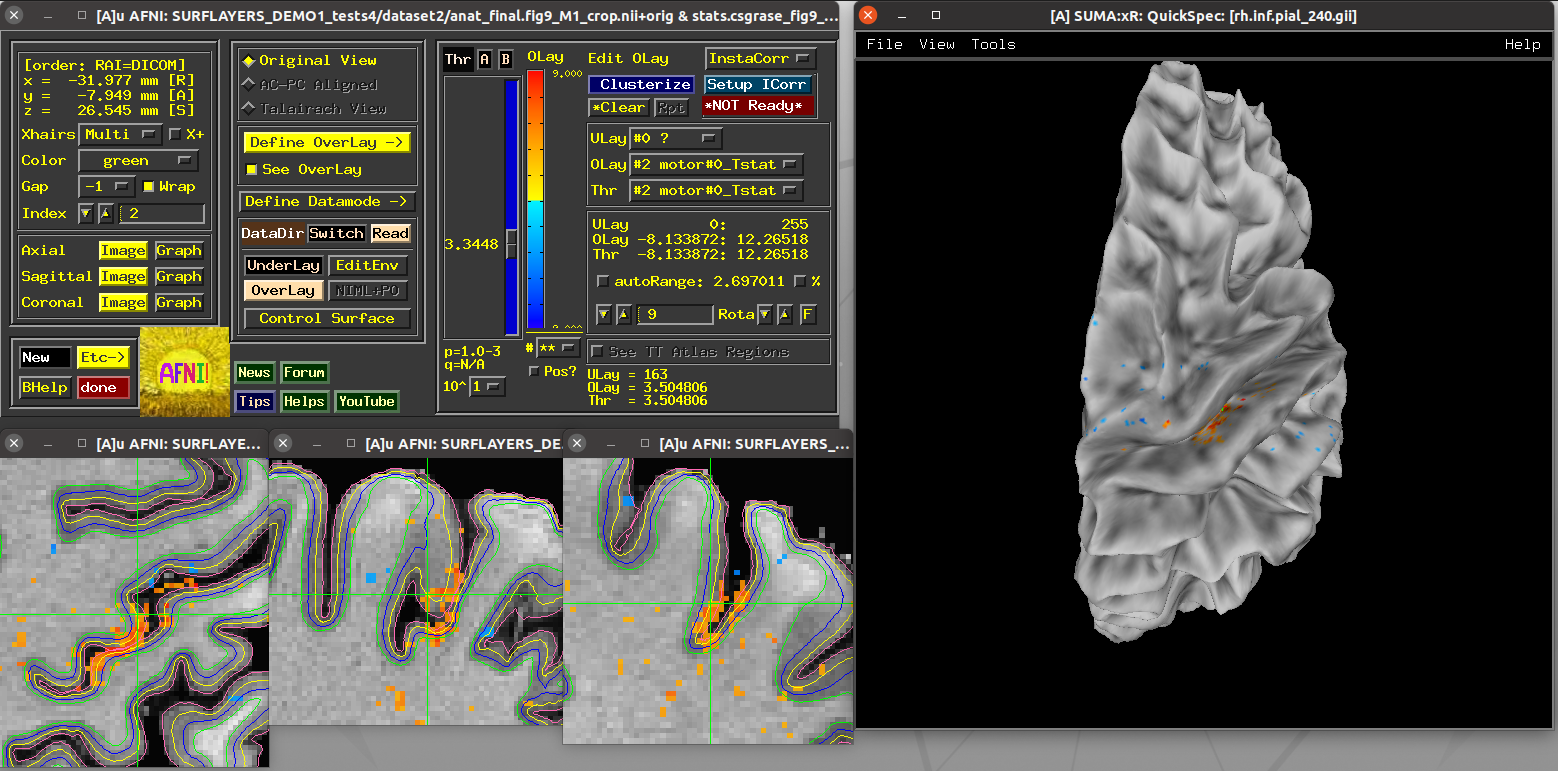
Running script |
|---|
|
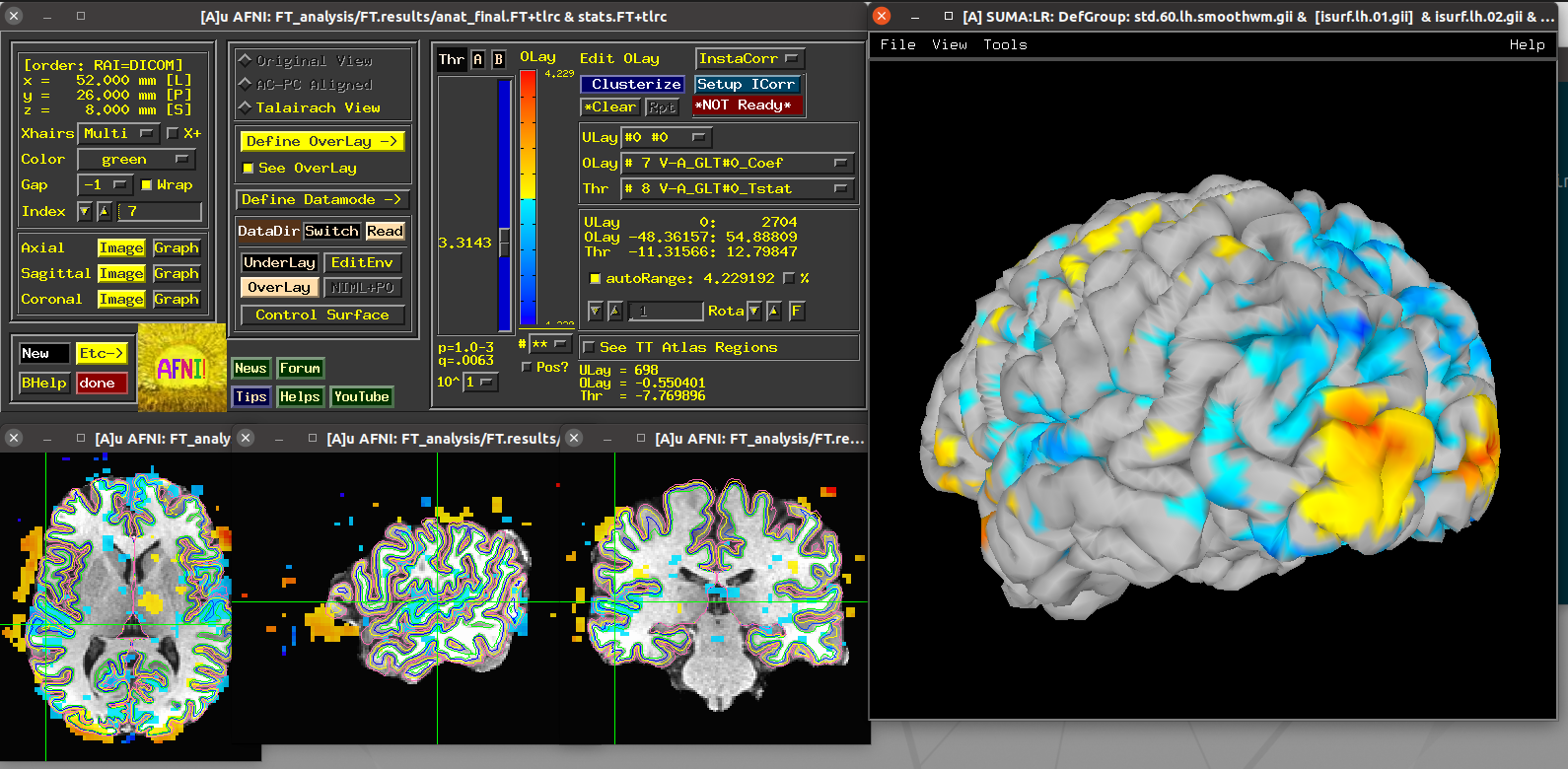
Scripts 3 and 4: laminar FMRI hemisphere¶
These scripts utilize data contained within the demo itself—and this
is a real, laminar FMRI dataset. The dataset2 directory contains
one anatomical (MP2RAGE) hemisphere and a 7T fingertapping dataset
using accelerated GRASE sequence.
Running script |
|---|
|
Running script |
|---|
|
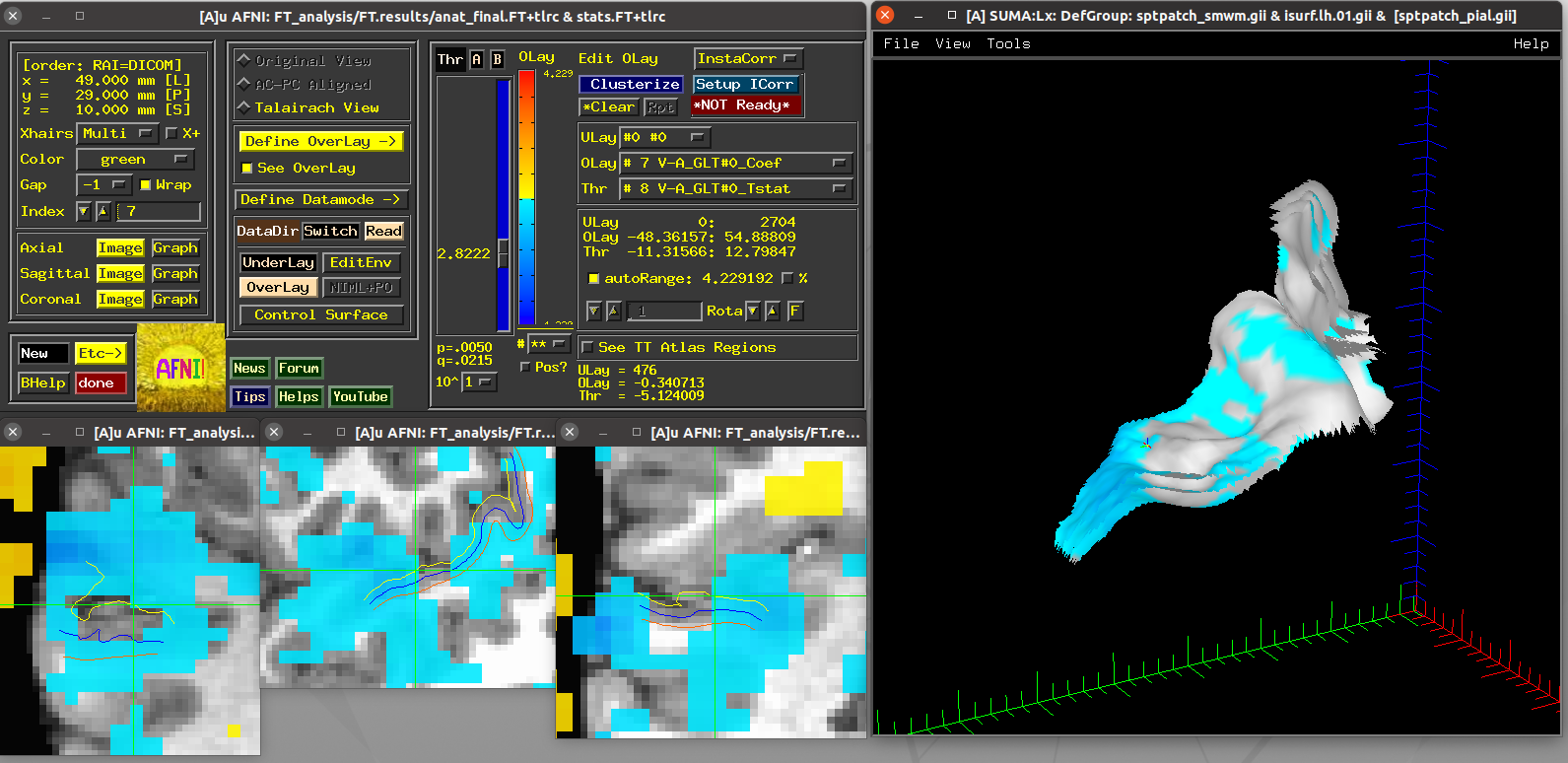
Scripts 5 and 6: laminar FMRI patch¶
These scripts also utilize laminar FMRI data contained within the demo
itself (also a 7T accelerated GRASE dataset with an MP2RAGE structural
scan). The dataset3 directory contains a left calcarine patch
that was drawn on retinotopic (meridian-mapping) functional results on
a spherical surface.
Running script |
|---|
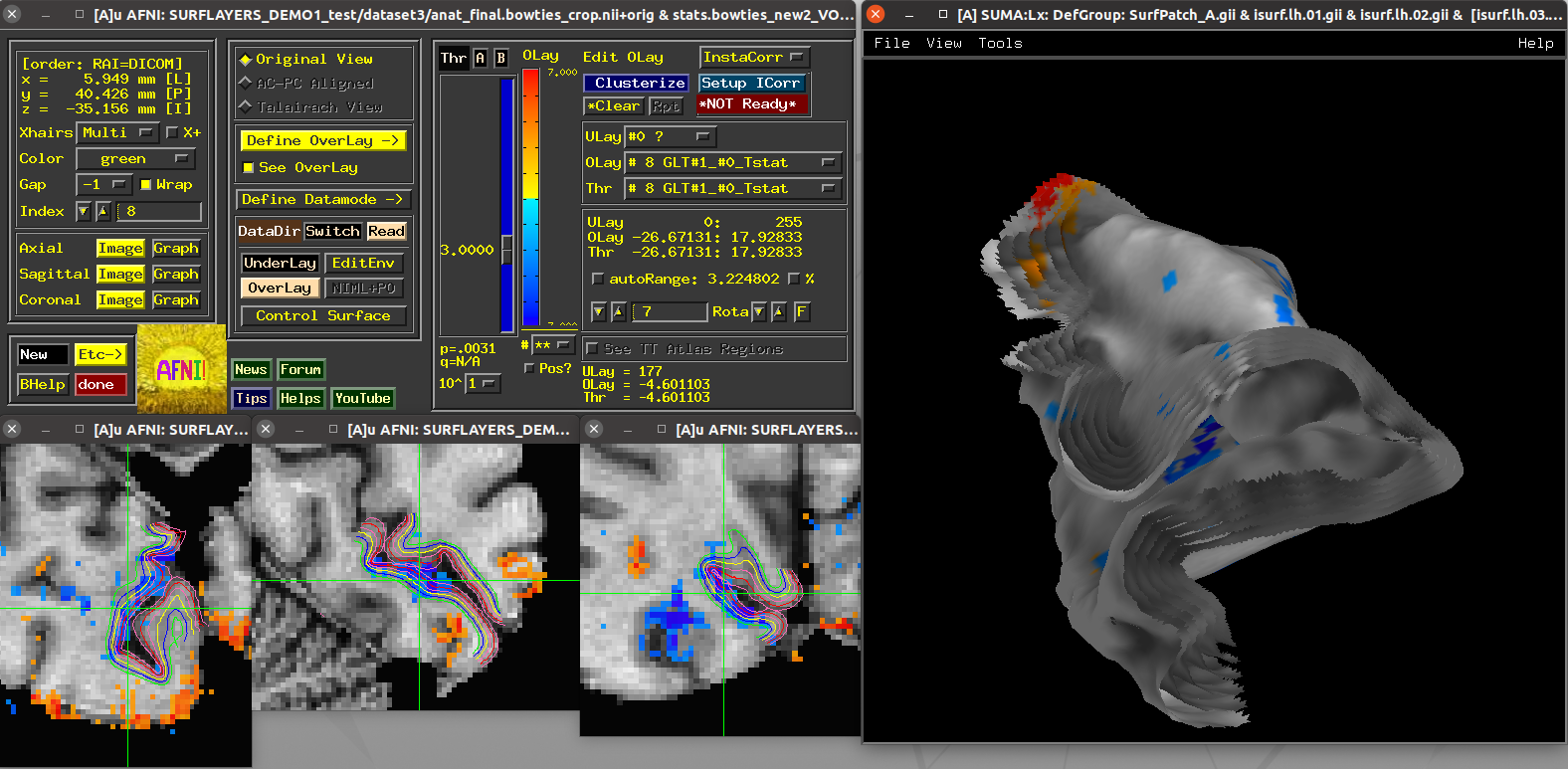
|
Running script |
|---|
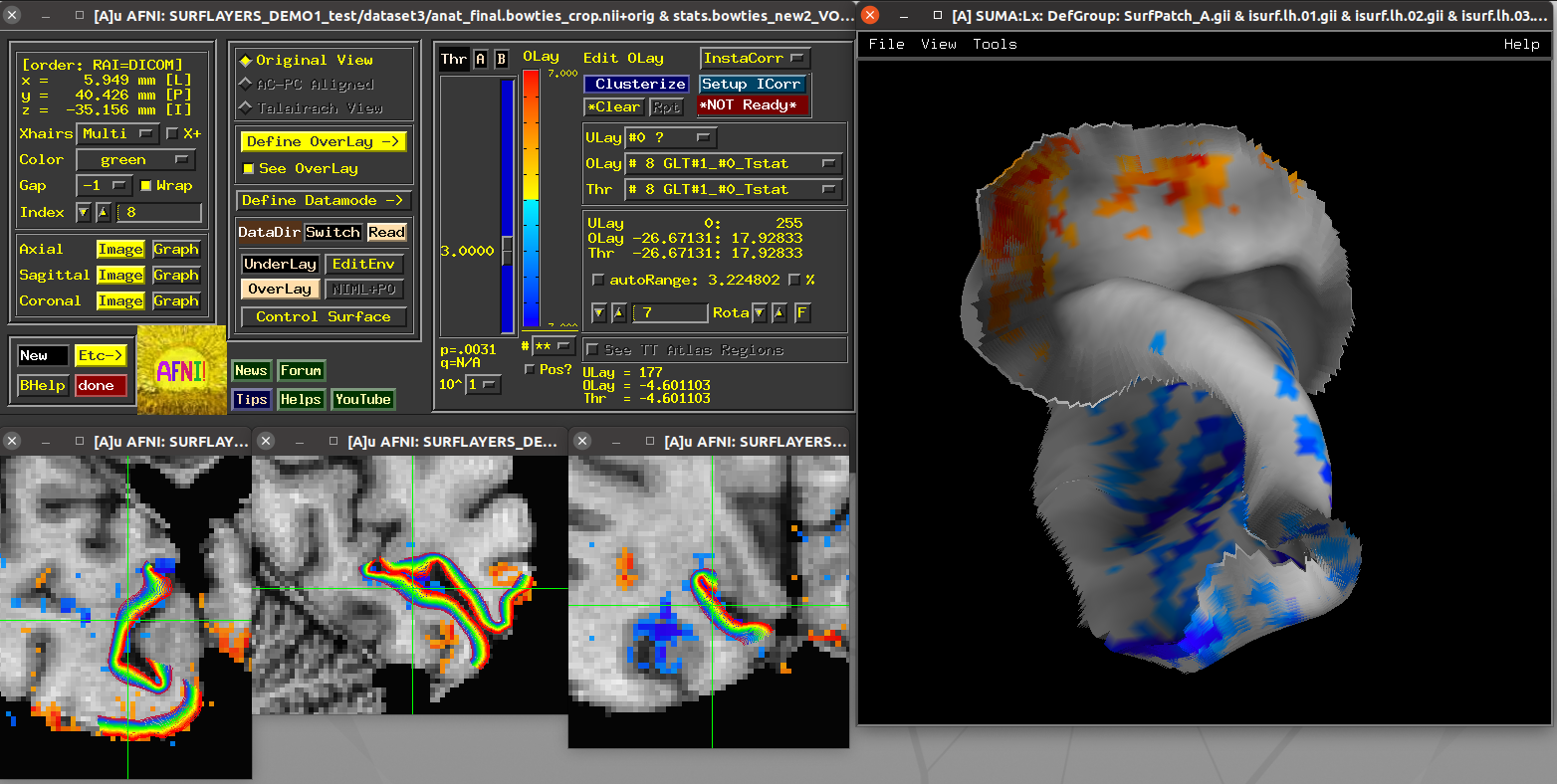
|
Questions/Contact¶
For questions/comments/suggestions, please contact:
Salvatore (Sam) Torrisi |
torrisi __at__ berkeley.edu |
Daniel Glen |
glend __at__ mail.nih.gov |
Paul Taylor |
paul.taylor __at__ nih.gov |
