Dealing with Obliquity in AFNI
For FMRI, oblique datasets are sometimes acquired for an EPI time
series dataset. An anatomical dataset is also often acquired in the
usual cardinal planes. For these datasets to correspond to each other
in AFNI, either the oblique EPI dataset can be transformed to a
cardinal orientation, or the cardinal anatomical dataset may be
transformed to match the oblique coordinates of the EPI dataset.
If DICOM data is read in with to3d, a matrix is computed for the transformation of the voxels as they are stored on the disk (i,j,k indices) to the real x,y,z coordinates in mm and stored in the dataset header information. Similarly, when NIFTI format datasets are used in AFNI, a transformation matrix is created in AFNI from the q-form or s-form fields.
There are several caveats however. The transformation changes the voxel data onto a new grid. In many cases, it may be preferable to transform the anatomical data to oblique instead of "deobliquing" the EPI time series dataset, but both options are available. When transforming EPI data to cardinal axes, the data should be time shifted with 3dTshift to account for the different acquisition times of each slice. The EPI data will, therefore, be interpolated in both time and space. The 3dWarp program handles both types of oblique transformations: oblique to cardinal and cardinal to oblique. The "-oblique2card" option effectively "deobliques" the oblique dataset (-deoblique is an alternative name for the same option, if that makes it clearer). Alternatively, the "-card2oblique" option requires an oblique parent dataset and transforms the cardinal dataset to oblique (-oblique_parent is an alternative name for this option). The new transformed datasets are computed on a new grid either specified by the -newgrid or -gridset options, or 3dWarp will assume the smallest spacing from the original dataset. This is an example script.
# read in the DICOM data
to3d -epan -assume_dicom_mosaic -prefix epiobliquet3 -time:zt 34 170 2 altplus EP/outslice.0*
# alternative 1 - deoblique EPI data
# first adjust for the slice timing before transforming EPI data
3dTshift -prefix epio_tshift epioblique+orig
3dWarp -oblique2card -prefix epio_card epio_tshift+orig
# alternative 2 - obliquify the anatomical data
3dWarp -card2oblique epiioblique+orig. -prefix anat_obl anat+orig.
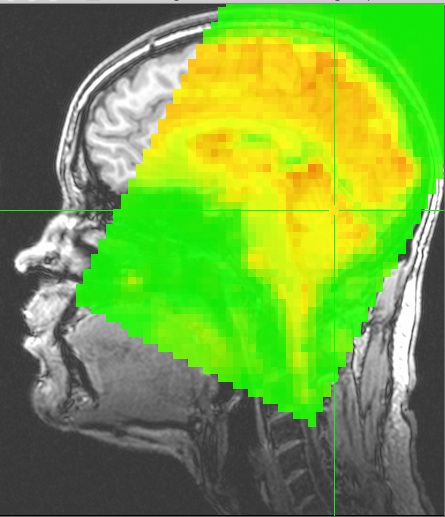
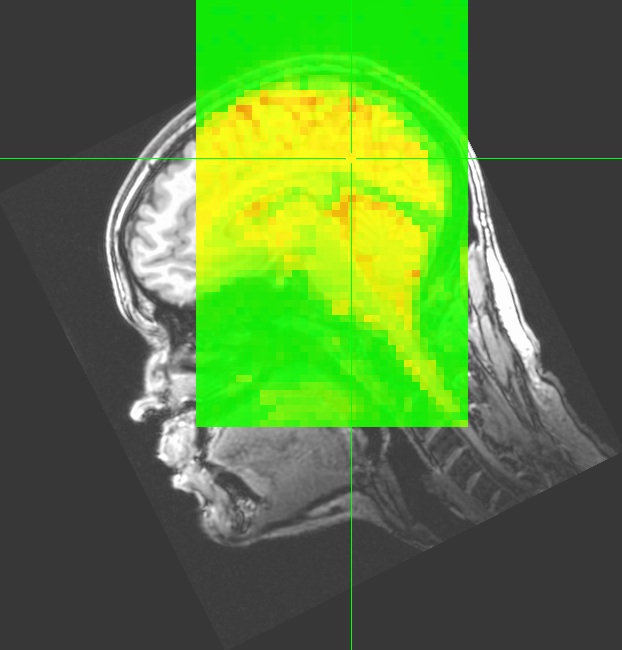
NIFTI datasets created by AFNI will carry the equivalent transformation information in the NIFTI header information in the qform and sform fields.
Questions and comments to Ziad Saad, Daniel Glen




