12.15.3. More useful outputs from @SUMA_Make_Spec_FS¶
Introduction¶
Download script: fs_makespec.tcsh
After running FreeSurfer’s (FS’s) recon-all, most AFNI users will
run @SUMA_Make_Spec_FS as described here or
here. This program has several jobs:
to convert FS volumetric and surface output to standard NIFTI and GIFTI datasets
to make standardized meshes from the estimated surfaces; see here for more details about how this is accomplished: Argall, Saad & Beauchamp (2006)
to derive new, additionally useful output files from the FS estimations.
This section is mainly dedicated to describing the 3rd category (derived information), which continues to grow over time.
The renumbered (REN) atlas dsets¶
@SUMA_Make_Spec_FS will call an AFNI program to take the standard
FS parcellation dsets, aparc+aseg.nii.gz (the “2000” atlas) and
aparc.a2009s+aseg.nii.gz (the “2009” atlas), and make renumbered
versions of the parcellations, named aparc+aseg_REN_*.nii.gz and
aparc.a2009s+aseg_REN_*.nii.gz, respectively. This is useful for
use with AFNI’s colorbars, and labeltables are also attached.
Additionally, there are several datasets made, grouped by tissue type.
Note
These tissue-grouped dsets are not binary, but contain the
renumbered ROI values. The categorizations are based on our
best guesses of where each ROI belongs, from both the
mri_binarize command in FS and our own supplementary
reading of the ROI names
The following files are output:
Dset suffix |
Description |
|---|---|
*_REN_gm.nii.gz |
gray matter |
*_REN_gmrois.nii.gz |
gray matter ROIs without *-Cerebral-Cortex ROI. This ROI file might be more useful for tracking or for making correlation matrices than *_REN_gm.nii.gz, because it doesn’t include the tiny scattered bits of the *-Cerebral-Cortex parcellation. |
*_REN_wmat.nii.gz |
white matter |
*_REN_csf.nii.gz |
cerebrospinal fluid |
*_REN_vent.nii.gz |
ventricles and choroid plexus |
*_REN_othr.nii.gz |
optic chiasm, non-WM-hypointens, etc. |
*_REN_unkn.nii.gz |
FS-defined “unknown”, with voxel value >0 |
The fs*.nii.gz mask dsets¶
There are a few masks that are created (mainly from the REN dsets, described above), typically named fs*nii.gz. These dsets are binarized (unlike the REN dsets, above):
fs_ap_wm.nii.gz: white matter mask, excluding the dotted part from FS. Useful for including in afni_proc.py for tissue-based regressors.
fs_ap_latvent.nii.gz: mask (not map!) of the lateral ventricles, ‘*-Lateral-Ventricle’. Useful for including in
afni_proc.pyfor tissue-based regressors in anaticor.fs_parc_wb_mask.nii.gz: a whole brain mask based on the FS parcellation. Note that this is different than the brainmask.nii* dset that FS creates. This mask is created in the following way:
binarize aparc+aseg_REN_all.nii.*
inflate by 2 voxels (3dmask_tool)
infill holes (3dmask_tool)
erode by 2 voxels (3dmask_tool)
The final mask seems much more specific to the brain structure than brainmask.nii* (image shown below). It also removes several small gaps and holes in the parcellation dset. In general, it seems like quite a useful whole brain mask.
QC images¶
Whenever performing any processing step, it is up to the human performing it to verify that things went OK. These automatically generated QC images help streamline this check, providing one convenient piece of QC: systematically made images at the whole brain, tissue and ROI levels.
Each is described here (with the same example anatomical used
throughout this FS+AFNI tutorial, in the Bootcamp directory
AFNI_data6/FT_analysis/FT/):
qc_00*.jpg: the overlay is the brainmask.nii* volume in red, and the subset of that volume that was parcellated by FS (in either the “2000” or “2009” atlases) is outlined in black. The idea for this formatting is that we do want to see the official FS brainmask, but we might also want to note its differences with the the binarized aparc+aseg file. We might prefer using one or the other dsets as a mask for other work.
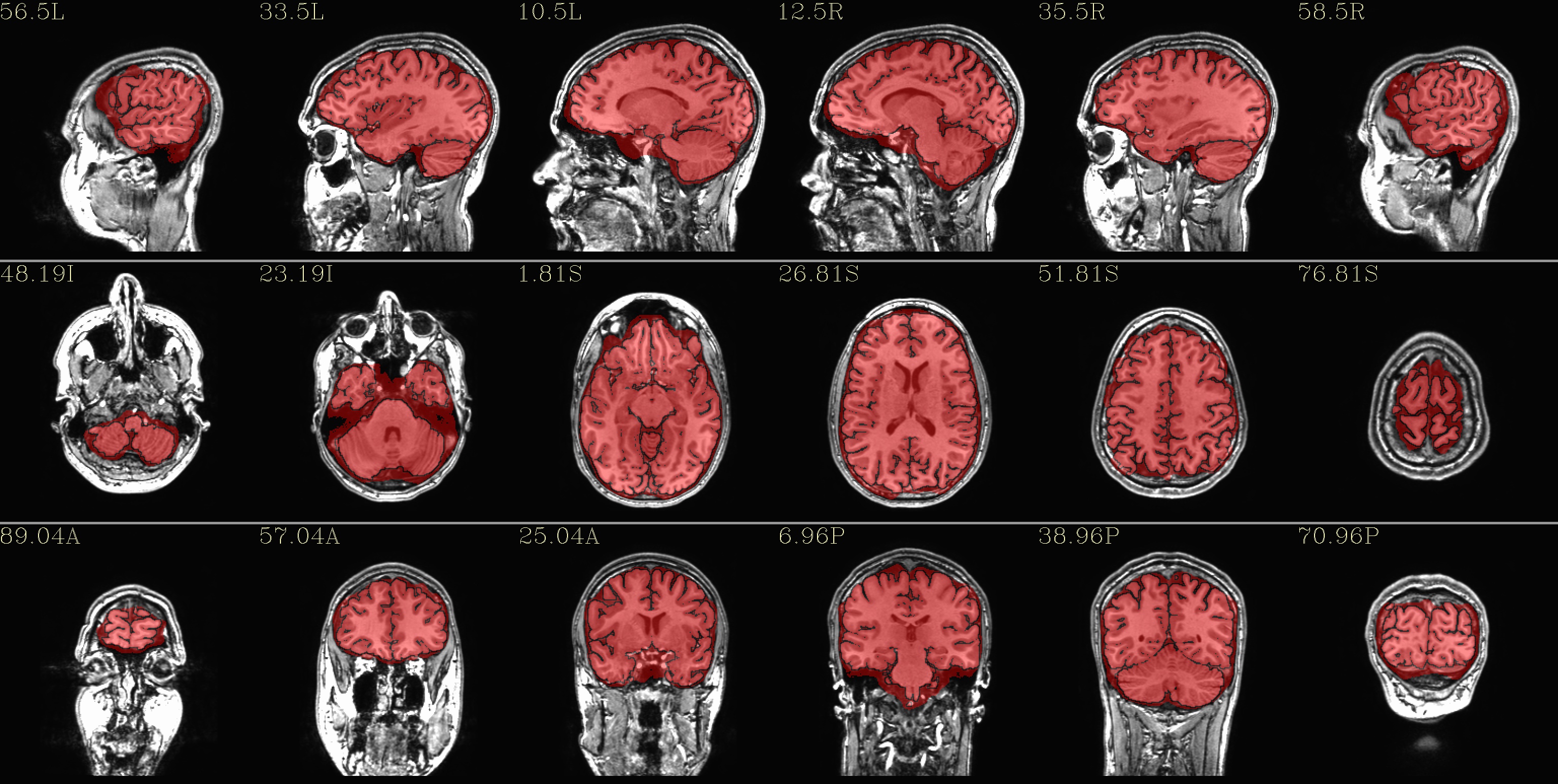
|
qc_01*.jpg: the overlay is the fs_parc_wb_mask.nii.gz dset that this script has created (see details just above).
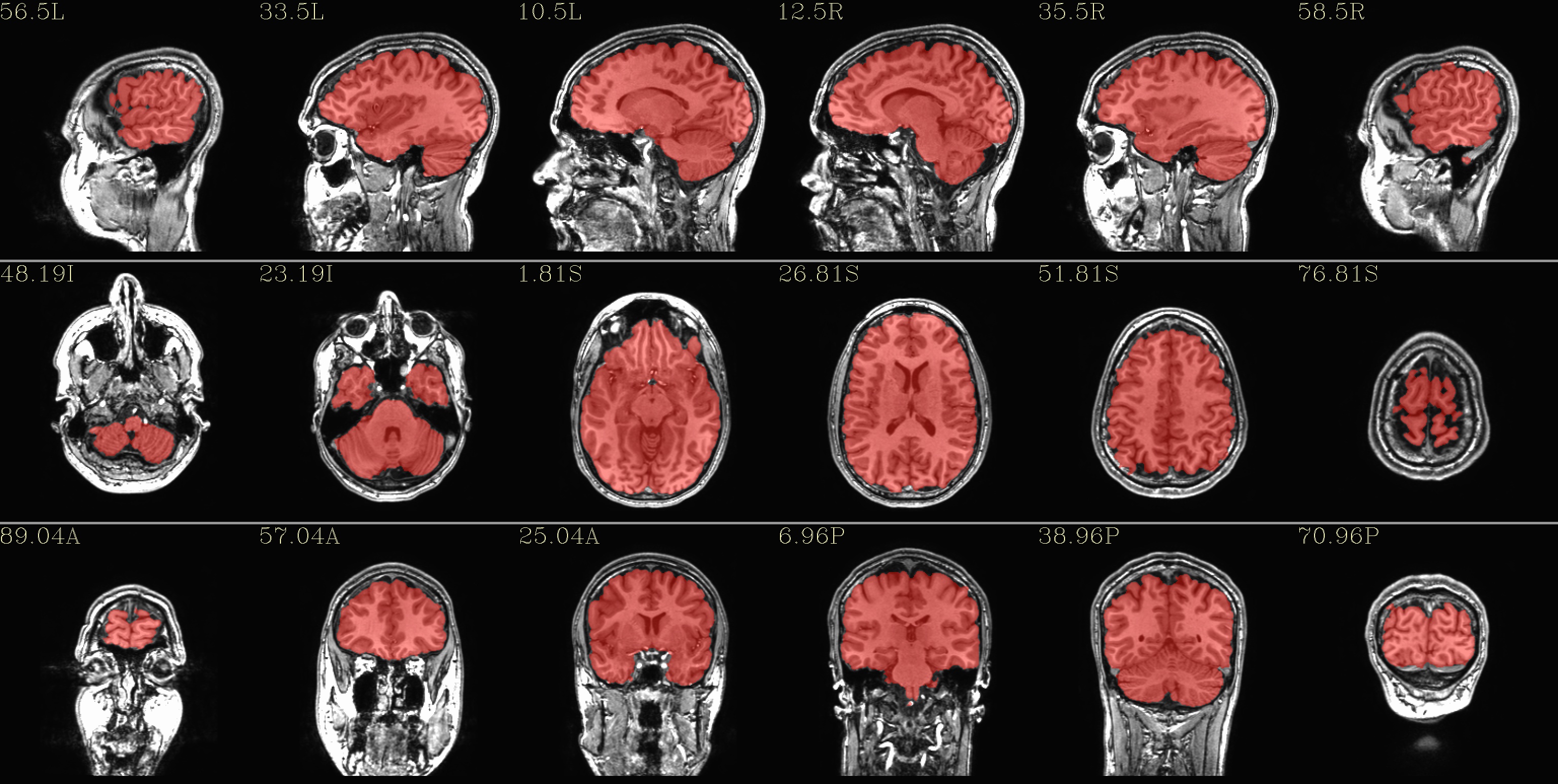
|
qc_02*.jpg: the overlay is a set of tissues, like a segmentation map of 4 classes:
red - GM - red
blue - WM
green - ventricles
violet - CSF+other+unknown
(from the REN files made by AFNI/SUMA).
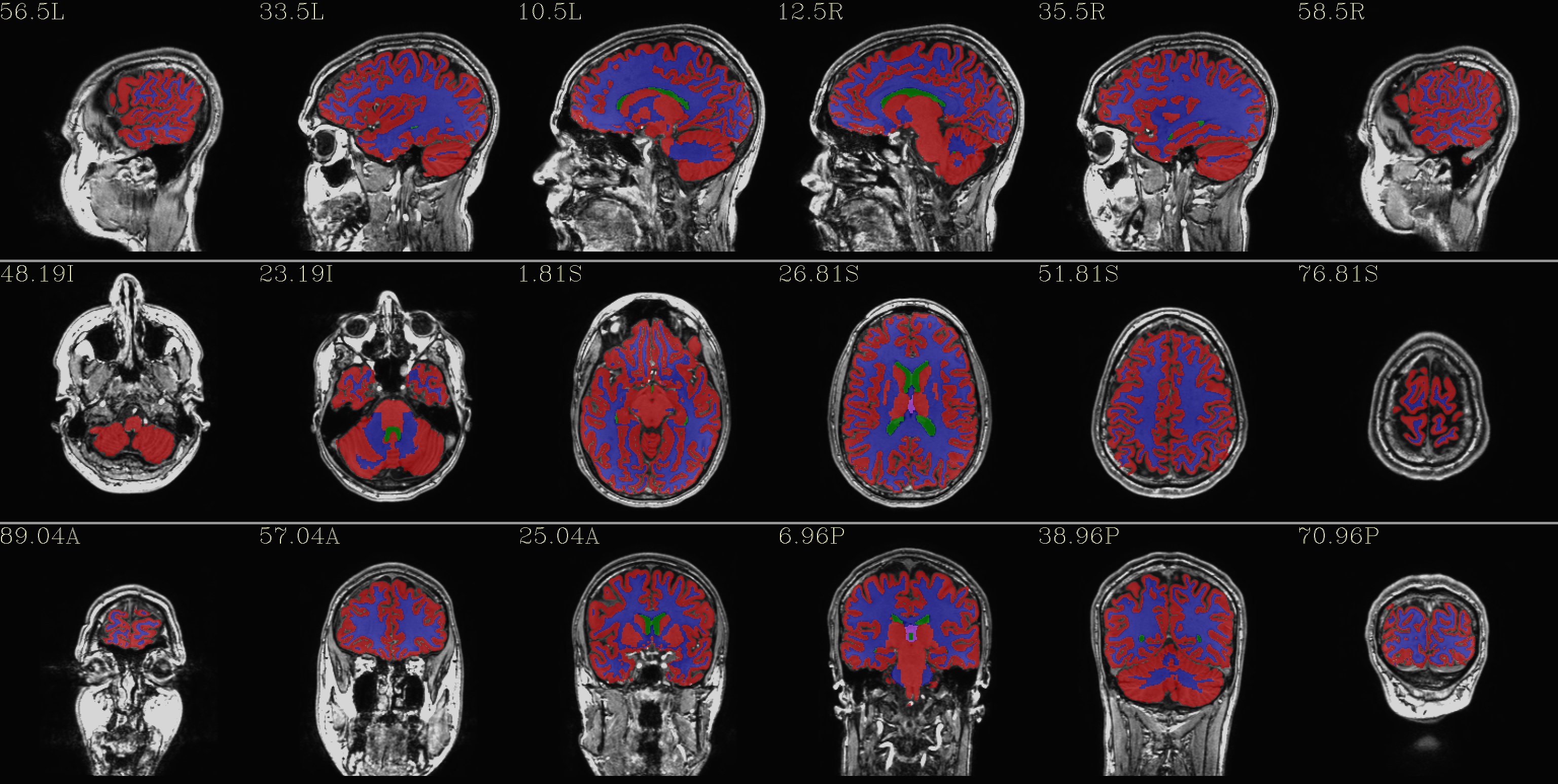
|
qc_03*.jpg: the GM only
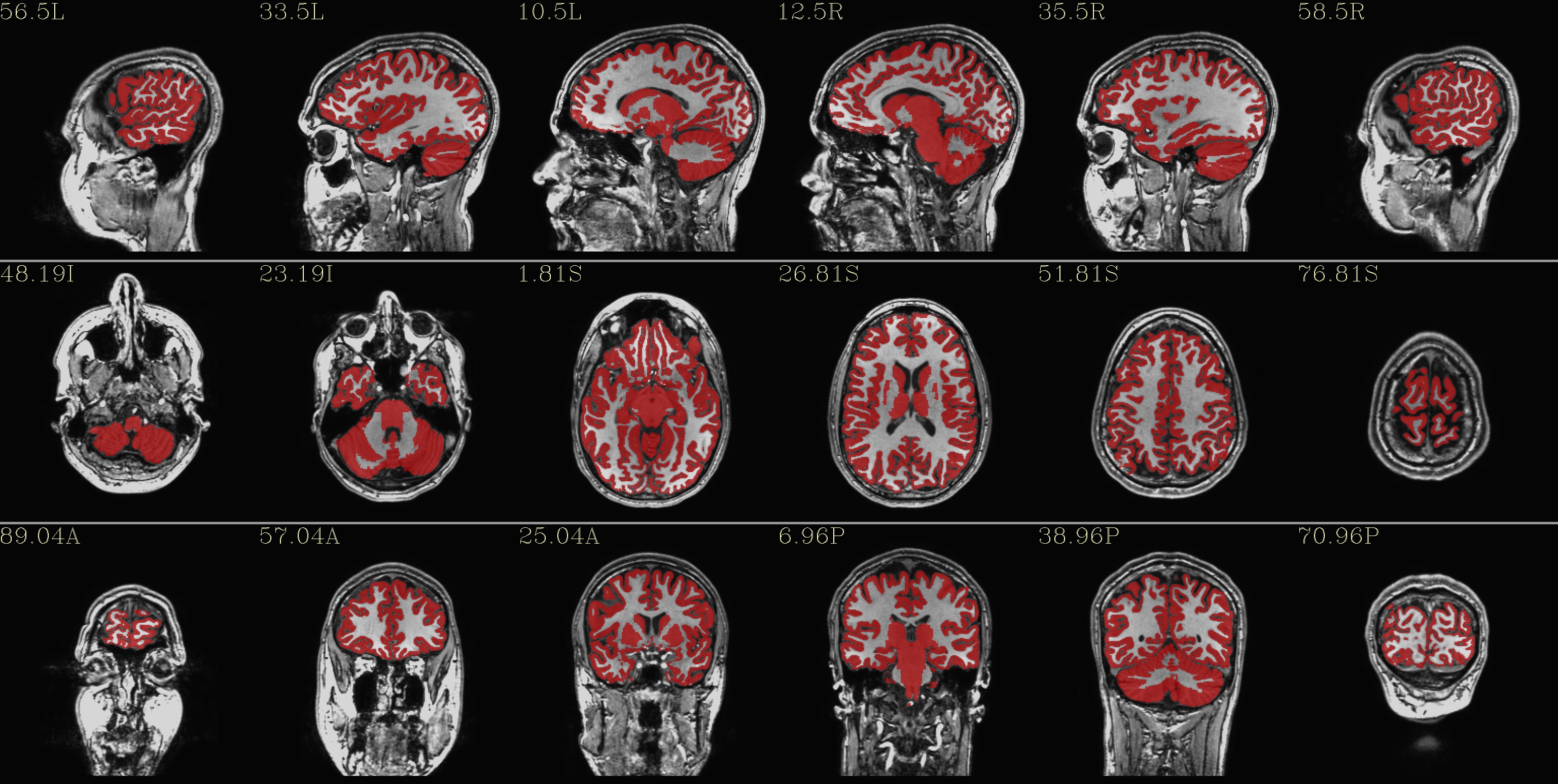
|
qc_04*.jpg: the WM only
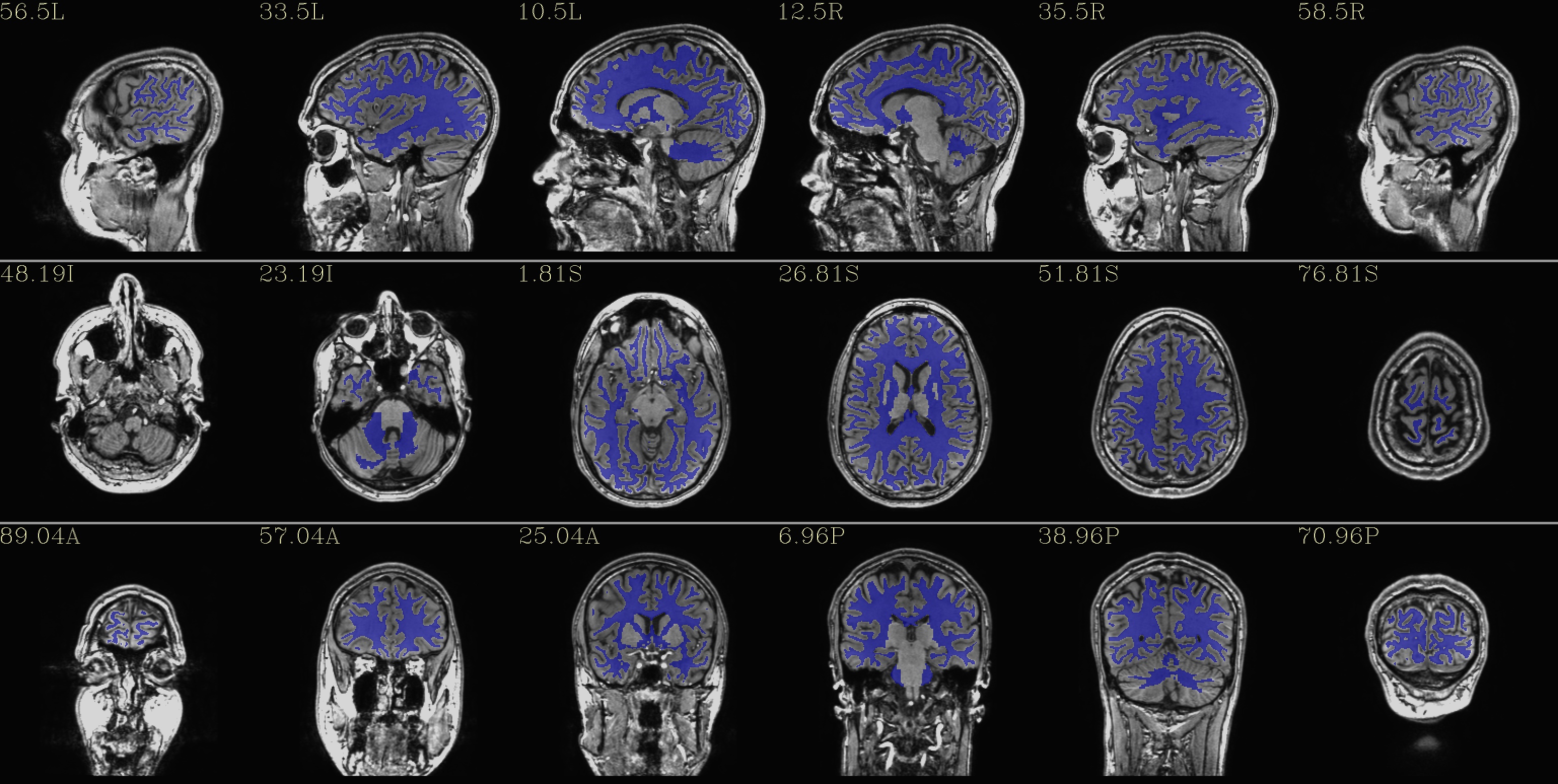
|
qc_05*.jpg: the overlay is the “2000” atlas parcellation (from the file: aparc+aseg*REN*all*)
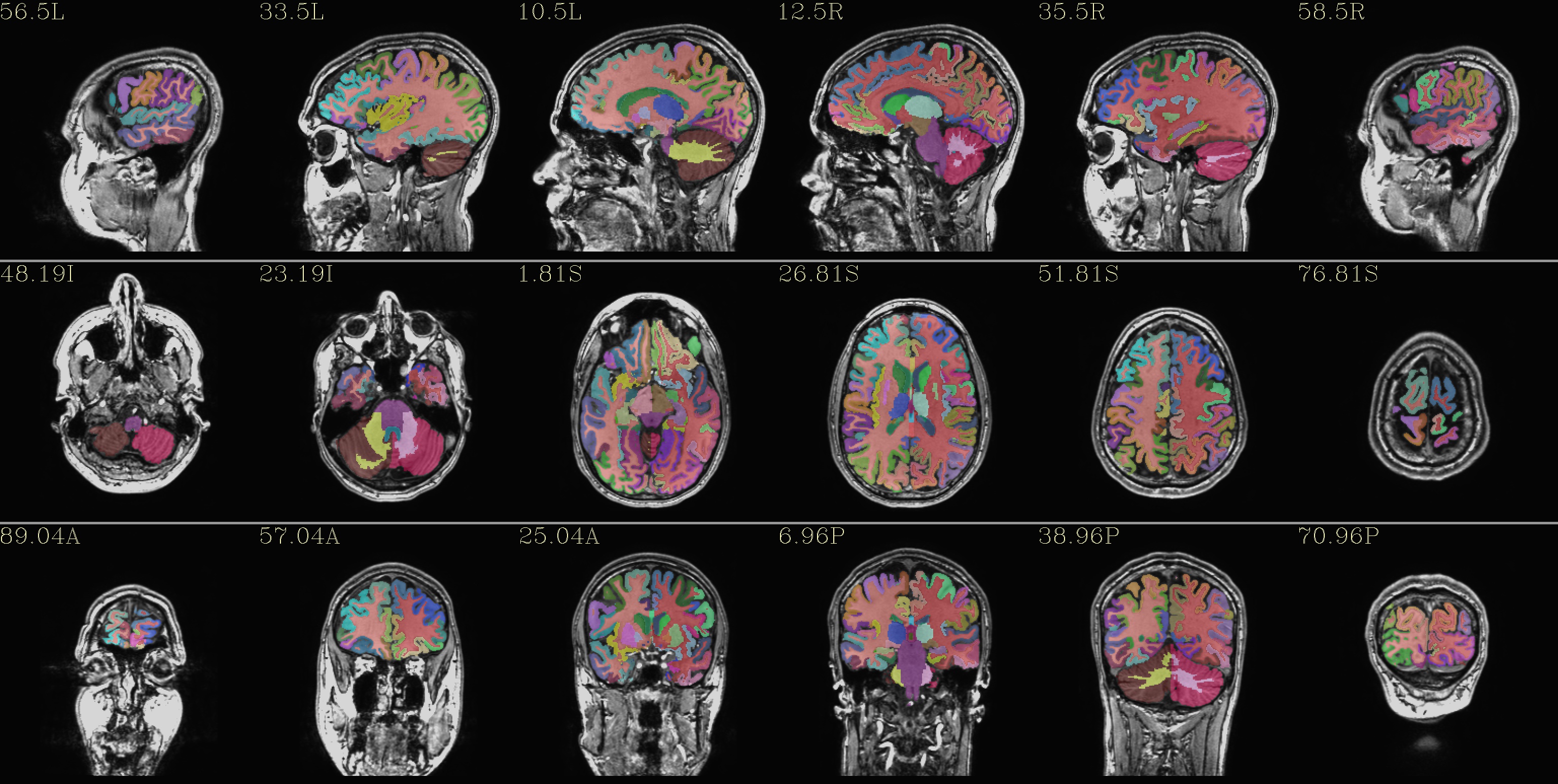
|
ROI, tissue and mask quantities (stats*.1D files)¶
Knowing the voxel count and relative fraction of various masks, tissue
maps and atlases can also be useful. That is why simple text files of
such relevant information are also created, in the stats*.1D files.
At present (and this might change over time as we think of more useful
things to calculate!), each file contains 4 columns of numbers,
followed by a comment symbol # and the ROI information. The
columns are:
Nvox: number of voxels in the ROI, segment or mask. This number is always an integer,
.
FR_BR_MASK: fraction of the number of voxels, segment or mask, relative to the “br_mask” dset (that is, to the brainmask.nii* volume).
FR_PARC_MASK: fraction of the number of voxels, segment or mask, relative to the “parc_mask” dset (that is, to the fs_parc_wb_mask.nii.gz volume that is created by the AFNI program adjunct_suma_fs_mask_and_qc). If this file does not exist, you will get a col of -1 values for the fraction; but you should just run adjunct_suma_fs_mask_and_qc.
fs_parc_wb_mask.nii.gz is a filled in form of the aparc+aseg segmentation result (see above).
FR_ALL_ROI: fraction of the number of voxels, segment or mask, relative to the full set of ROIs in the given parcellation (that is, to the *REN_all.nii* volume).
And here are examples of the first few lines of these info files for the “2000” atlas, and similar exists for the “2009” atlas.
For stats_fs_segs_2000_*.1D, describing the REN datasets and masks:
# Nvox FR_BR_MASK FR_PARC_MASK FR_ALL_ROI # SEG__TYPE FILE_NAME
1904388 1.000000 1.197520 1.316392 # br_mask brainmask.nii
1590277 0.835059 1.000000 1.099266 # parc_mask fs_parc_wb_mask.nii.gz
1446672 0.759652 0.909698 1.000000 # all aparc+aseg_REN_all.nii.gz
849797 0.446231 0.534370 0.587415 # gm aparc+aseg_REN_gm.nii.gz
849797 0.446231 0.534370 0.587415 # gmrois aparc+aseg_REN_gmrois.nii.gz
575808 0.302359 0.362080 0.398022 # wmat aparc+aseg_REN_wmat.nii.gz
19628 0.010307 0.012343 0.013568 # vent aparc+aseg_REN_vent.nii.gz
1382 0.000726 0.000869 0.000955 # csf aparc+aseg_REN_csf.nii.gz
57 0.000030 0.000036 0.000039 # othr aparc+aseg_REN_othr.nii.gz
For stats_fs_rois_2000_*.1D, describing the ROI parcellations:
# Nvox FR_BR_MASK FR_PARC_MASK FR_ALL_ROI # VAL TISS__TYPE STRING_LABEL
0 0.000000 0.000000 0.000000 # 0 tiss__unkn Unknown
271443 0.142535 0.170689 0.187633 # 1 tiss__wmat Left-Cerebral-White-Matter
0 0.000000 0.000000 0.000000 # 2 tiss__gm Left-Cerebral-Cortex
6917 0.003632 0.004349 0.004781 # 3 tiss__vent Left-Lateral-Ventricle
150 0.000079 0.000094 0.000104 # 4 tiss__vent Left-Inf-Lat-Vent
15797 0.008295 0.009933 0.010920 # 5 tiss__wmat Left-Cerebellum-White-Matter
65932 0.034621 0.041459 0.045575 # 6 tiss__gm Left-Cerebellum-Cortex
10300 0.005408 0.006477 0.007120 # 7 tiss__gm Left-Thalamus-Proper
4311 0.002264 0.002711 0.002980 # 8 tiss__gm Left-Caudate
