12.12.5. Using 2dcat¶
Introduction¶
Download script tarball: auto_2dcat.tgz
Open the tarball using tar -xf auto_2dcat.tgz to get the following scripts in your directory: tut_auto_2dcat_0.tcsh, tut_auto_2dcat_1.tcsh, tut_auto_2dcat_2.tcsh.
Download script: tut_auto_2dcat_0.tcsh
2dcat is an “image concatenation” program. This is a very useful
supplementary tool for glueing existing images together to make arrays
with separating lines, as well other features. It combines quite
usefully with @chauffeur_afni, for example to concatenate similar
images across a data set.
We present some examples of auto-image making with 2dcat using
data that should be available in (modern) AFNI binary distributions,
the *_SSW.nii.gz template targets for @SSwarper. Each of
these dsets has multiple bricks. We provide examples:
combining multiple views and slices of the same dataset
adjoining similar views across separate datasets
and more.
Each short script creates a subdirectory (“QC_2dcat*”) for the both the individual, intermediate images and the final concatenated matrix of images.
Ex. 0: Combine images subject- and slice-wise¶
Variables have been defined so that one should be able to adapt these scripts by changing the just file name(s) in the uppermost input section.
Definitions at top of script:
- show code y/n -Use @chauffeur_afni to make individual images¶
Each @chauffeur_afni execution creates a set of sagittal, axial
and coronal images; each image output by chauffeur here is an 8x1
montage. These will later be glued together.
set allbase = ()
foreach ff ( $ilist )
set ibase = `3dinfo -prefix_noext "${ff}"` # base name of vol
set allbase = ( $allbase $ibase ) # list of all base names
# Make a montage of the zeroth brick of each dset
@chauffeur_afni \
-ulay "${ff}[0]" \
-prefix ${odir}/img0_${ibase} \
-set_dicom_xyz 5 18 18 \
-delta_slices 10 20 10 \
-set_xhairs OFF \
-montx 8 -monty 1 \
-label_mode 1 -label_size 3 \
-do_clean
end
Use 2dcat to concatenate sliceviews for each subj¶
First example of using 2dcat on a set of datasets: for each dset,
concatenate different slice views (sagittal, coronal and axial) of a
single volume.
This example requires having the *_SSW.nii.gz template targets
copied into the present working directory. Alternatively, one could
just include the path to them in the glob at the top of the script
(e.g., set ivol = `\ls ~/abin/*${istr}.nii.gz`)
# Just the "gap color" between glued-together images
set lcol = ( 66 184 254 )
# For each volume, concatenate images across all 3 sliceviews. The
# order of contanenation will be that of globbing; could be specified
# in different ways, too.
foreach ff ( $allbase )
2dcat \
-gap 5 \
-gap_col ${lcol} \
-nx 1 \
-ny 3 \
-prefix $odir/ALL_subj_${ff}.jpg \
${odir}/img0_*${ff}*
end
Combined sliceviews for each subject |
|---|
HaskinsPeds_NL_template1.0_SSW: |

|
MNI152_2009_template_SSW: |
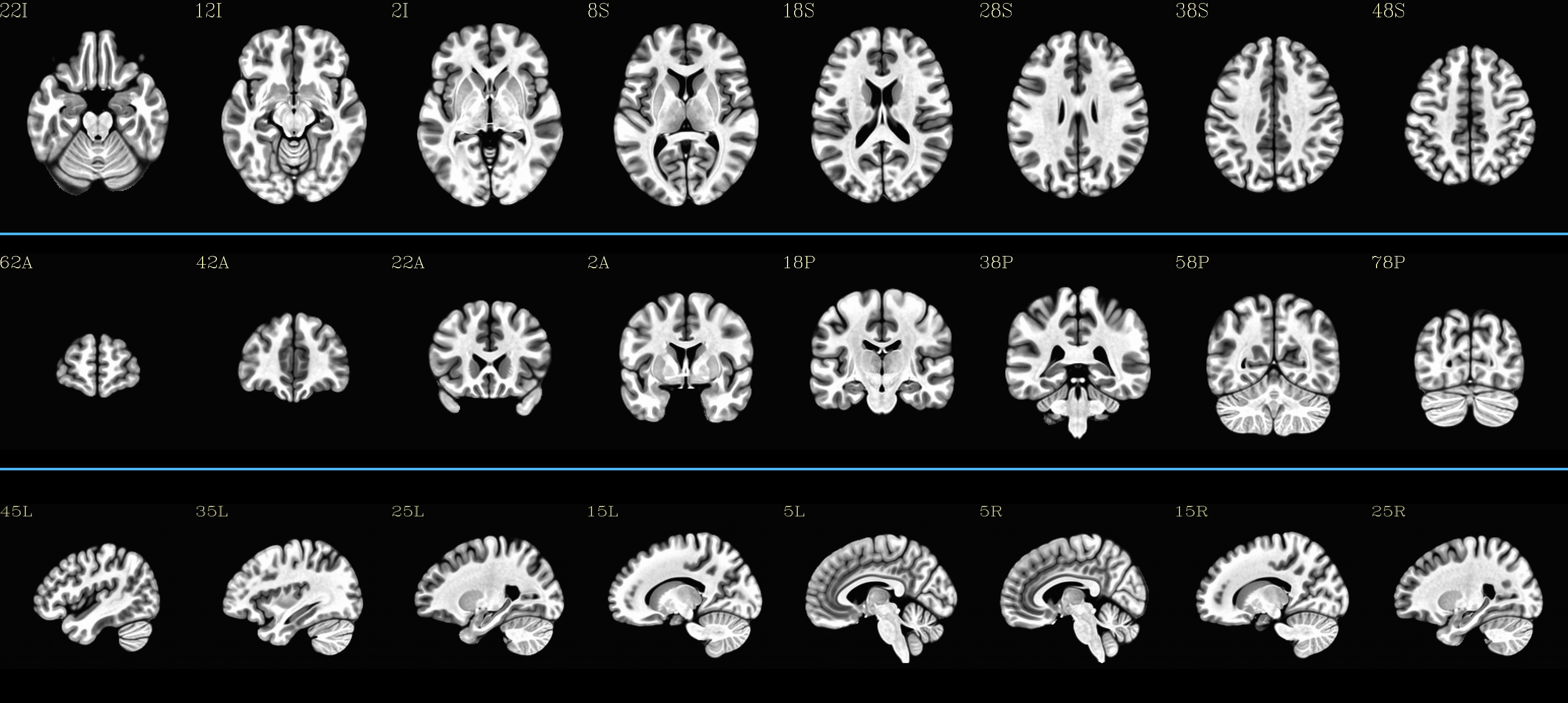
|
TT_N27_SSW: |

|
Use 2dcat to concatenate subjs for each sliceview¶
Second example of using 2dcat on a set of datasets: for each slice
view, show the dset at the same (x, y, z) location.
# Just the "gap color" between glued-together images
set lcol = ( 255 152 11 )
# For each sliceview, concatenate images across all vols
foreach ss ( "sag" "cor" "axi" )
2dcat \
-gap 5 \
-gap_col ${lcol} \
-nx 1 \
-ny ${Ndset} \
-prefix $odir/ALL_${istr}_sview_${ss}.jpg \
${odir}/img0_*${ss}*
end
# ---------------------------------------------------------------------
echo "++ DONE!"
# All fine
exit 0
Combined subjects for each sliceview |
|---|
sagittal views: |

|
coronal views: |
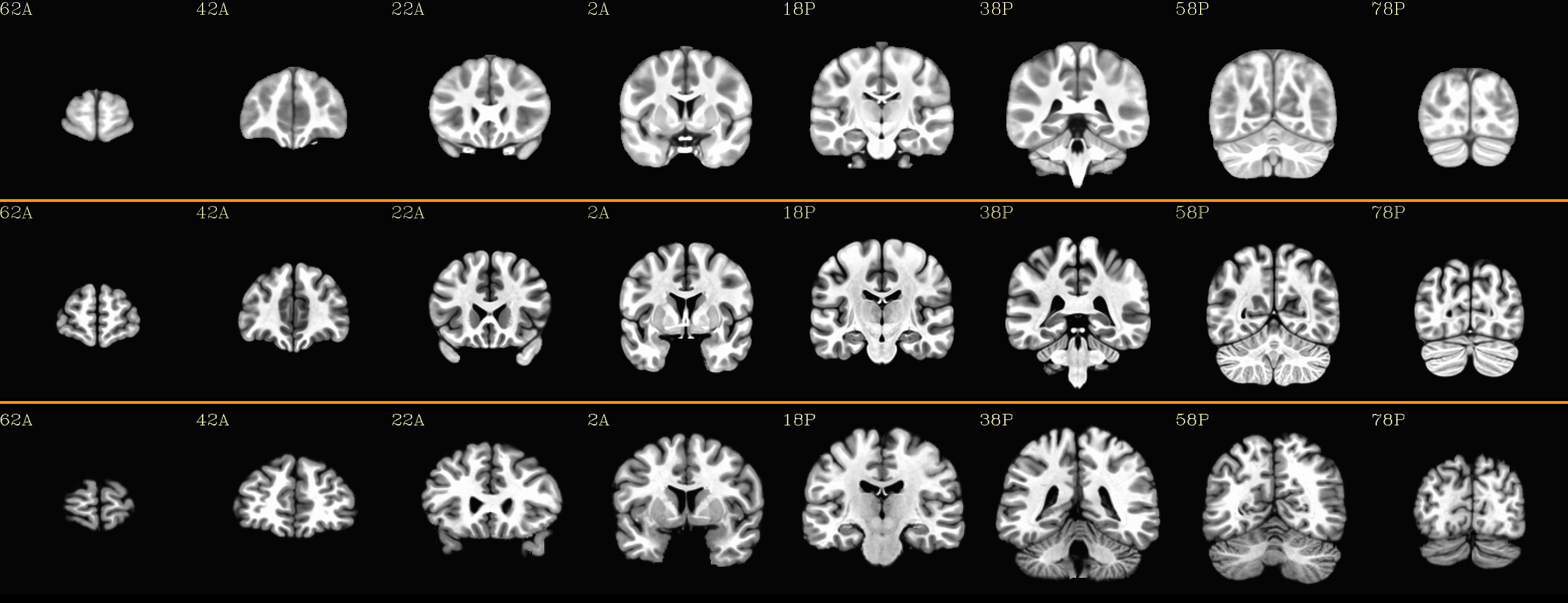
|
axial views: |
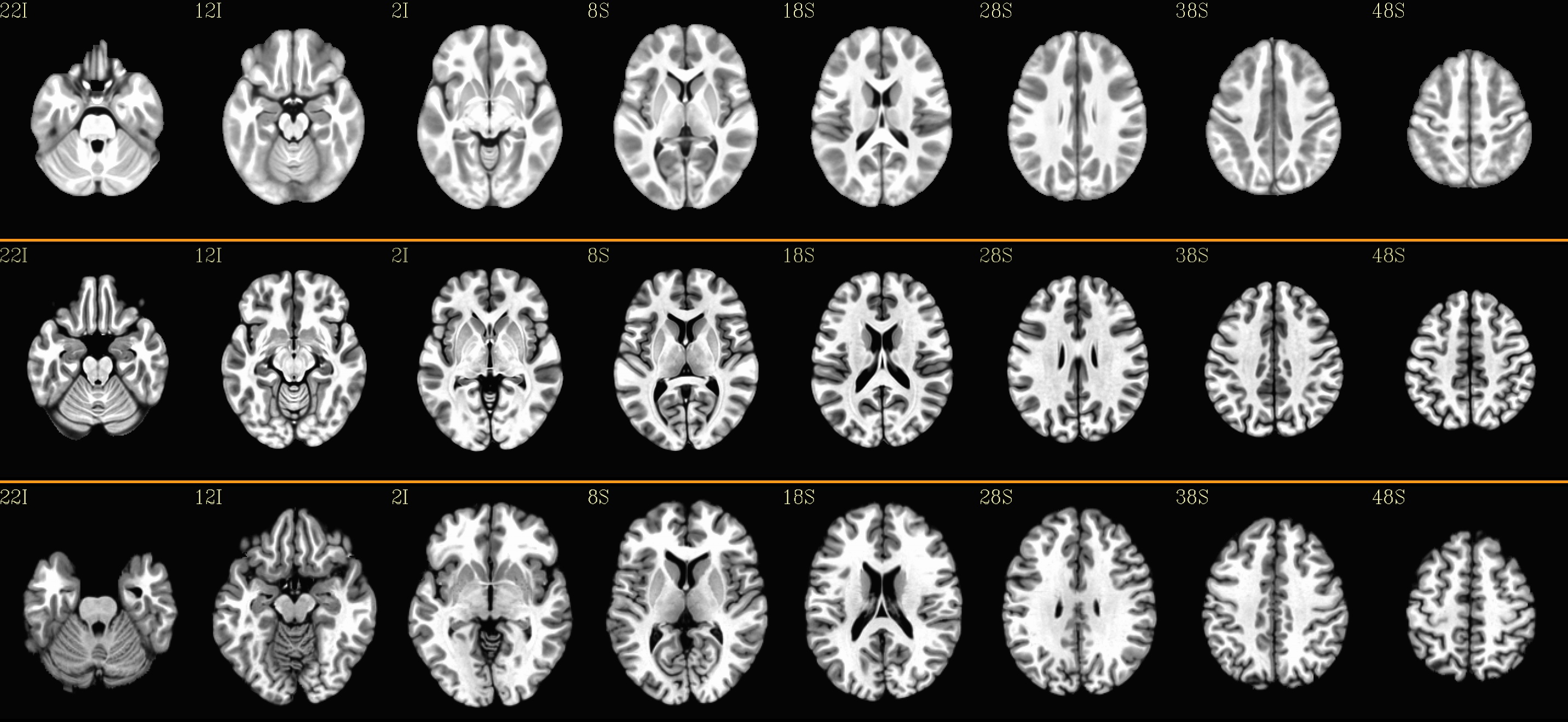
|
Ex. 1: Combine subbrick images of a 4D dset¶
Download script: tut_auto_2dcat_1.tcsh
Make a set of sagittal, axial and coronal images; these will later be glued together. Here, we are make a set of images per volume in a 4D dset.
Definitions at top of script:
- show code y/n -Use @chauffeur_afni to make individual images¶
foreach ii ( `seq 0 1 ${imax}` )
# zeropadded numbers, nicer to use in case we have a lot of images
set iii = `printf "%03d" ${ii}`
# This if-condition is a sidestep: we have two categories of data
# in the input volume, masks and dsets, with very different
# pertinent ranges, so we account for that here.
if ( ${ii} > 2 ) then
set UMIN = "0"
set UMAX = "1"
else
set UMIN = "2%"
set UMAX = "98%"
endif
@chauffeur_afni \
-ulay "${ivol}[$ii]" \
-ulay_range "$UMIN" "$UMAX" \
-prefix ${odir}/${ibase}_${iii} \
-set_dicom_xyz 2 18 18 \
-delta_slices 25 25 25 \
-set_xhairs OFF \
-montx 1 -monty 1 \
-label_mode 1 -label_size 3 \
-do_clean
end
Use 2dcat to concatenate images¶
Combine the individual images from above into a matrix of images. Here we have three rows (i.e., three images along y-axis: one for sagittal, axial and coronal), and the number of columns is equal to the number of volumes in the 4D dset.
# concatenate 3 sliceviews, for as many volumes as are in the dset
2dcat \
-echo_edu \
-gap 5 \
-gap_col ${lcol} \
-nx ${nv} \
-ny 3 \
-prefix $odir/ALL_vol_${ibase}.jpg \
$odir/${ibase}*sag* $odir/${ibase}*cor* $odir/${ibase}*axi*
# ---------------------------------------------------------------------
echo "++ DONE!"
# All fine
exit 0
Ex. 1: Each subject & all sliceviews |
|---|
MNI152_2009_template_SSW: |

|
Ex. 2: Combine (stats) images from many subj¶
Download script: tut_auto_2dcat_2.tcsh
Here we present a nice way to make a summary of similar images across a group of subjects. In this case, we use a set of individual modeling results: we threshold based on a statistical criterion (voxelwise p<0.001, two-sided) and show the effect estimates (beta coefficients).
We can apply the typical hard thresholding, where everything in subthreshold voxels is hidden. Or, we can use a more modern alpha thresholding, whereby subthreshold voxels are merely made increasingly transparent as their values are further below threshold.
Definitions at top of script:
- show code y/n -Use @chauffeur_afni to make individual images¶
set allbase = ()
foreach ff ( ${ilist} )
# base name of vol, and make a list of all prefixes for later
set ibase = `3dinfo -prefix_noext "${ff}"`
set allbase = ( ${allbase} ${ibase} )
### Make a montage of the zeroth brick of each image.
# Some fun-ness here: part of each file's name is added to the
# label string shown in each panel.
# Note: these olay datasets are unclustered and unmasked.
@chauffeur_afni \
-ulay ${ianat} \
-ulay_range "2%" "130%" \
-olay ${ff} \
-set_subbricks -1 0 1 \
-func_range 3 \
-thr_olay_p2stat 0.001 \
-thr_olay_pside bisided \
-cbar Reds_and_Blues_Inv \
-olay_alpha Yes \
-olay_boxed Yes \
-opacity 7 \
-prefix ${odir}/img0_alpha_${ibase} \
-montx 1 -monty 1 \
-set_dicom_xyz 5 18 18 \
-set_xhairs OFF \
-label_string "::${ibase}" \
-label_mode 1 -label_size 3 \
-do_clean
# same images as above, but with hard thresholding
@chauffeur_afni \
-ulay ${ianat} \
-ulay_range "2%" "130%" \
-olay ${ff} \
-set_subbricks -1 0 1 \
-func_range 3 \
-thr_olay_p2stat 0.001 \
-thr_olay_pside bisided \
-cbar Reds_and_Blues_Inv \
-opacity 7 \
-prefix ${odir}/img0_hthr_${ibase} \
-montx 1 -monty 1 \
-set_dicom_xyz 5 18 18 \
-set_xhairs OFF \
-label_string "::${ibase}" \
-label_mode 1 -label_size 3 \
-do_clean
end
Use 2dcat to concatenate images¶
Combine the individual images from above into a matrix of images. Here we combine similar slice views. Note how we end up having a nice summary of subject modeling results across the group.
Scripting note : Note that here the nx and ny values are
hardcoded in, but they needn’t be, so this script could be more
flexible to match adding/subtracting subjects. Fancier things can be
done– feel free to ask/discuss/recommend suggestions.
foreach ss ( "sag" "cor" "axi" )
# Combine alpha-thresholded images
2dcat \
-echo_edu \
-gap 5 \
-gap_col ${lcol} \
-nx 5 \
-ny 2 \
-prefix ${odir}/ALL_alpha_${istr}_sview_${ss}.jpg \
${odir}/img0_alpha*${ss}*
# Combine hard-thresholded images
2dcat \
-echo_edu \
-gap 5 \
-gap_col ${lcol} \
-nx 5 \
-ny 2 \
-prefix ${odir}/ALL_hthr_${istr}_sview_${ss}.jpg \
${odir}/img0_hthr_*${ss}*
end
# ---------------------------------------------------------------------
echo "++ DONE!"
# All fine
exit 0
Ex. 2: One stat slice across subjects: alpha+boxed thresholding |
|---|
sagittal views: |
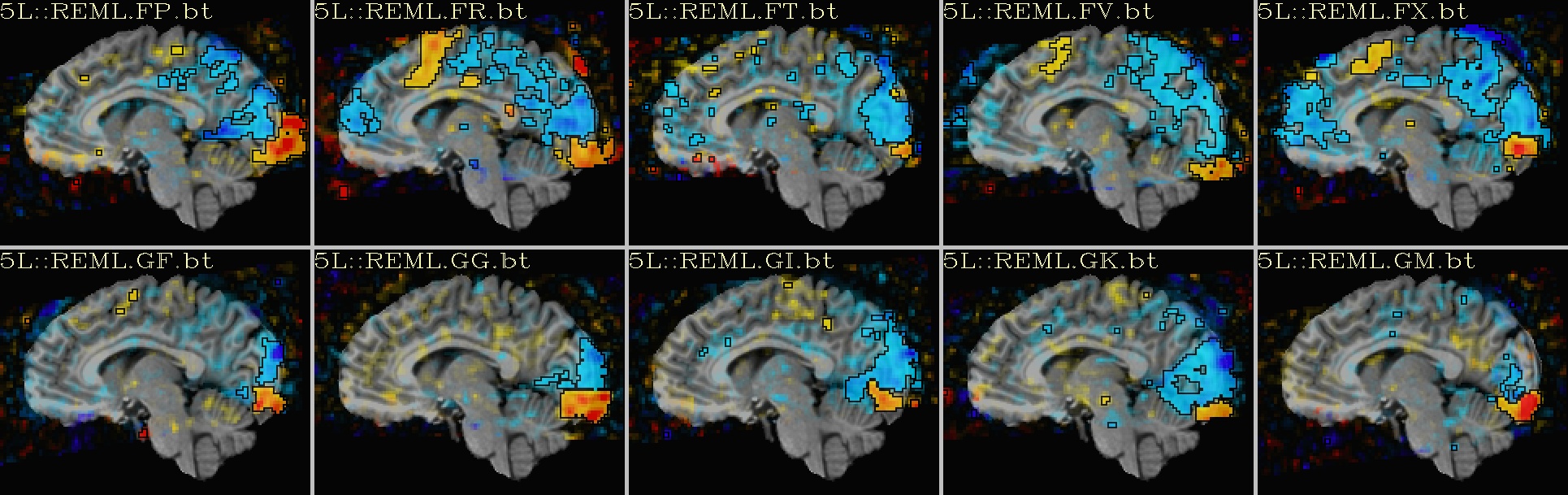
|
coronal views: |

|
axial views: |

|
Ex. 2: One stat slice across subjects: hard thresholding |
|---|
sagittal views: |

|
coronal views: |
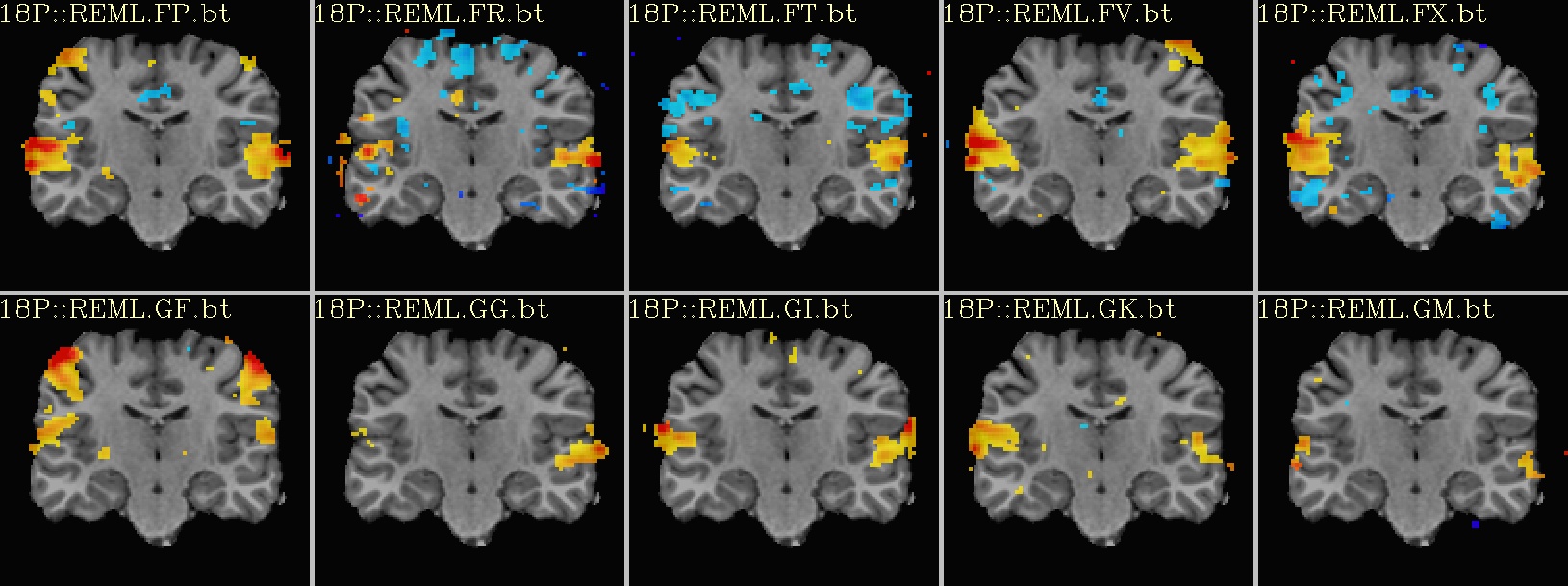
|
axial views: |
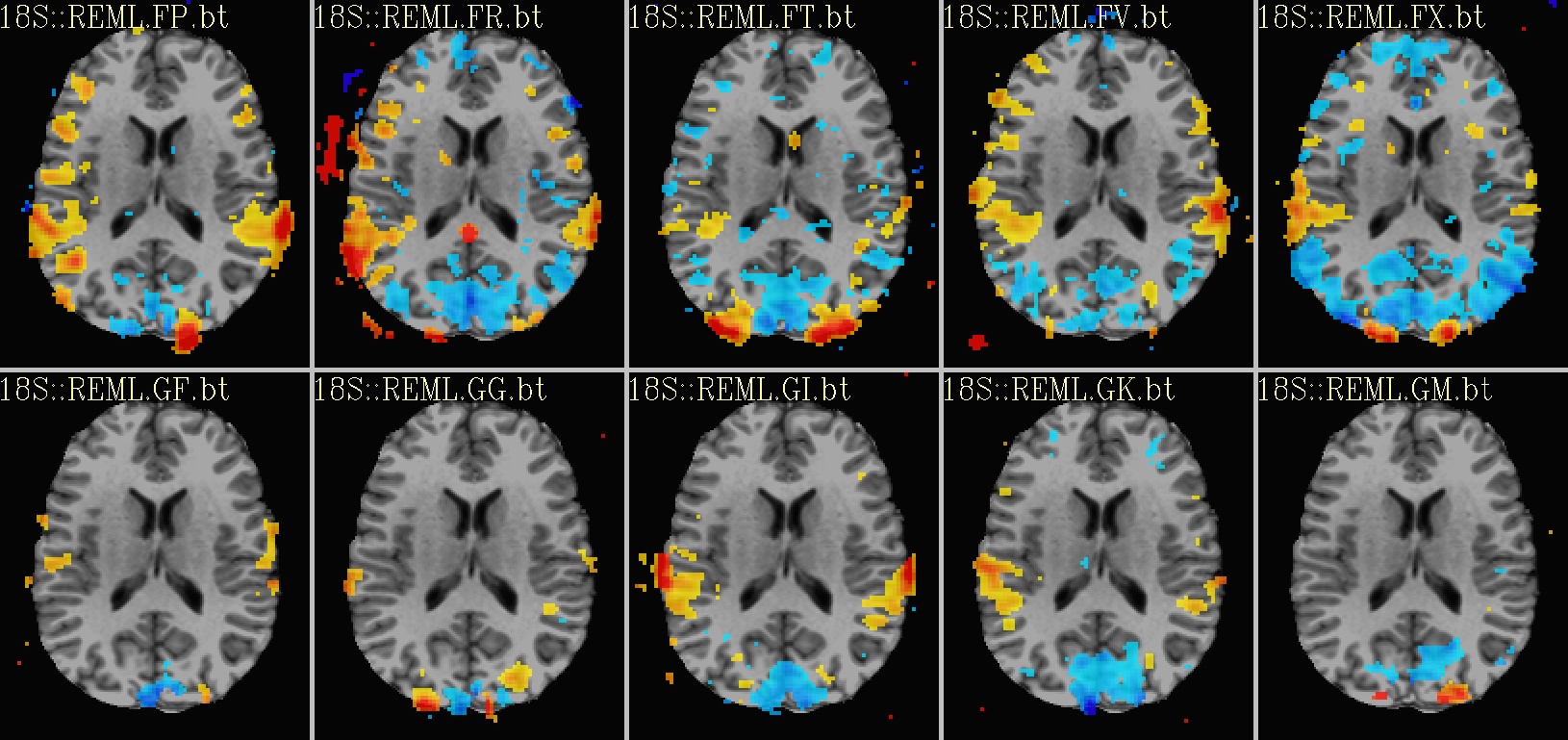
|
